Protein Folding
We are using cellular automata (CA) for the modeling of the temporal folding of proteins [2][3][6]. Unlike the focus of the vast research already done on the direct prediction of the final folded conformations [1][5], we model the temporal and dynamic folding process. The CA model defines how the amino acids interact through time to obtain a folded conformation. We employed the HP model to represent the protein conformations in a lattice, using evolutionary computing to automatically obtain the models.
-
Example of different temporal steps in the folding process with protein sequence HPPHPPHPPHPPHPPHPPHPPH
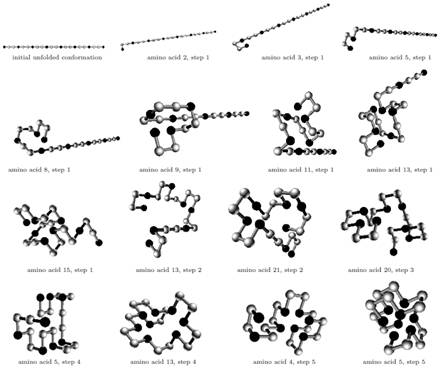
-
Animated sequences: Folding process with a protein in a 2D HP lattice and a protein in a 3D HP lattice:

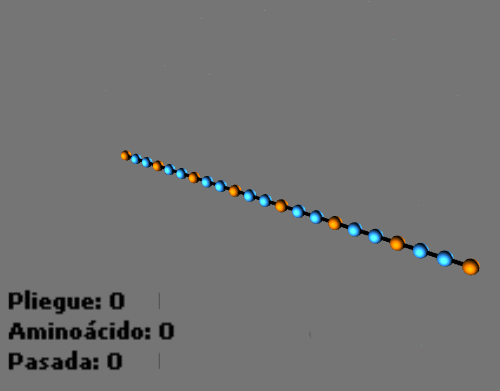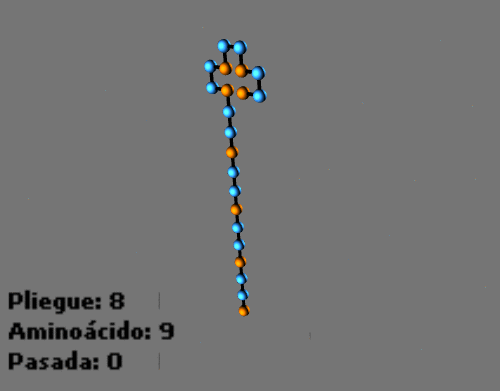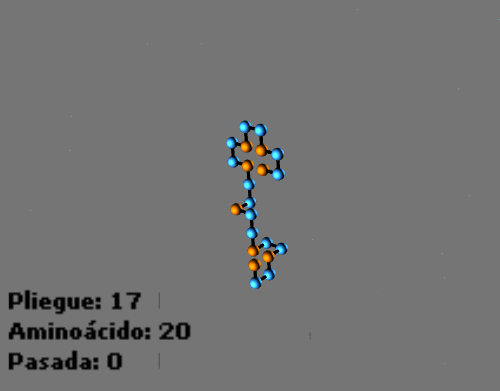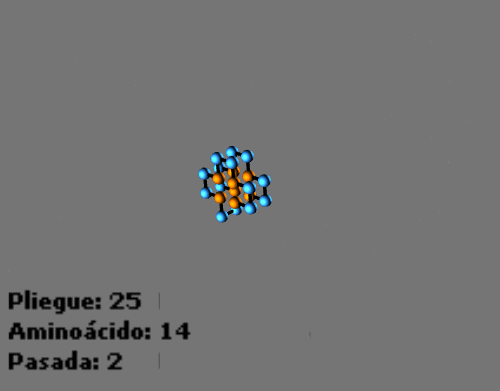
-
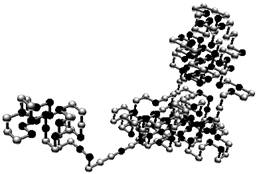
Final folding, using the HP model, with protein 3no6 from PDB (Protein Data Bank).
-
Folding example of a protein using the FCC lattice model:
-
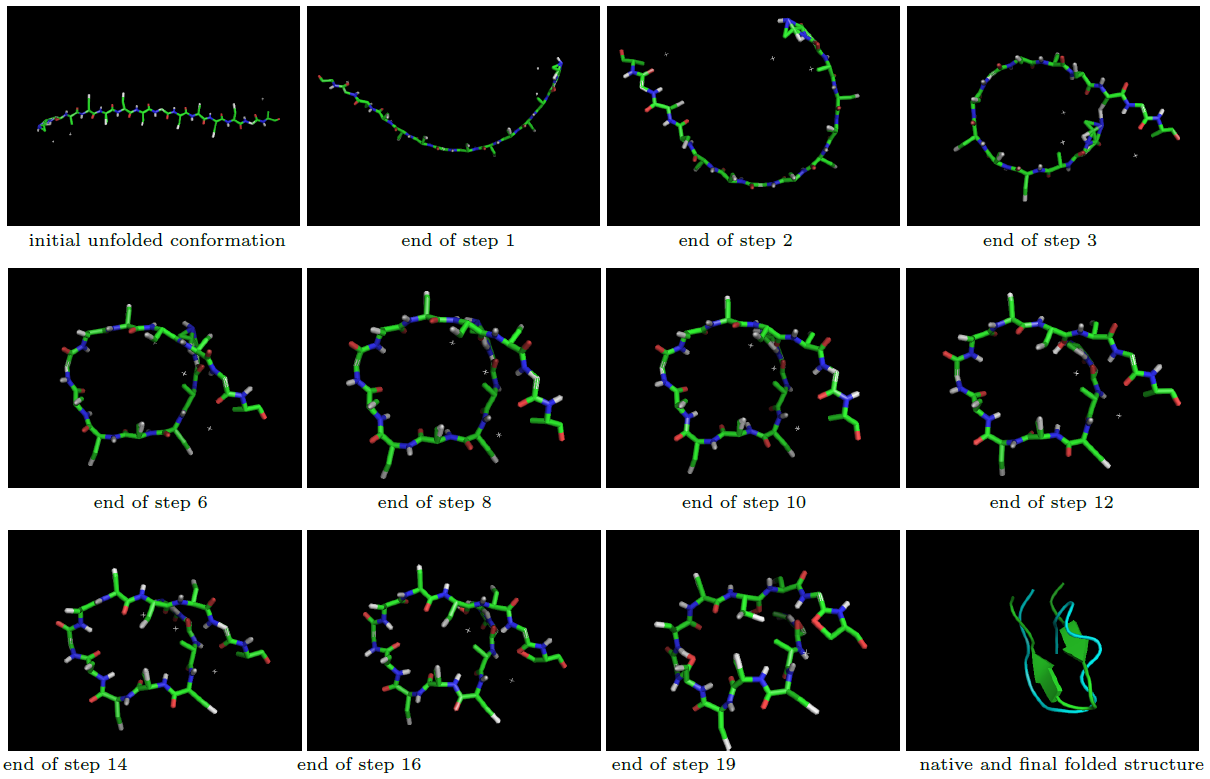
Snapshots of a folding example using the off-lattice coarse-grain representation model of Rosetta,with protein sequence 1j4m. The last snapshot shows the native structure (green) and the final folded structure at the last step (blue):
References with more information
[1] Santos. J. and Diéguez, M. (2011), Differential evolution for protein structure prediction using the HP model, Inter. Work-Conf. on the Interplay between Natural and Artificial Comp., LNCS 6686:323-333.
[2] Santos, J., Villot, P., Diéguez, M. (2013), Cellular automata for modeling protein folding using the HP model, Proc. IEEE Congress on Evolutionary Comp. - IEEE-CEC 2013, 1586-1593.
[3] Santos, J., Villot, P., Diéguez, M. (2013), Protein folding with cellular automata in the 3D HP model, Proc. Intern. Workshop Evolutionary Comp. in Bioinformatics - Genetic and Evol. Comp. Conf. (GECCO 2013), 1595-1602.
[4] Santos, J., Villot, P., Diéguez, M. (2014), Emergent protein folding modeled with evolved neural cellular automata using the 3D HP model, Journal of Computational Biology 21(11):823-845.
[5] Varela, D. and Santos, J. (2016), Protein folding modeling with neural cellular automata using Rosetta, GECCO 2016 Proceedings Companion, Workshop Evolutionary Computation in Computational Structural Biology, 1307- 1312.
[6] Varela, D. and Santos, J. (2015), Combination of differential evolution and fragment-based replacements for protein structure prediction GECCO 2015 Proceedings Companion, Workshop Evolutionary Computation in Computational Structural Biology, 911-914.
[7] Zhao, X. (2008), Advances on protein folding simulations based on the lattice HP models with natural computing, Applied Soft Computing 8:1029-1040.


