The genetic code
The canonical genetic code is not universal although it is present in most complex genomes. Its establishment is still under discussion once the discovery of non-standard genetic codes altered the frozen accident [1]. Since the canonical code is not universal, different theories about its origin and organization have appeared. The optimization or level of adaptation of the canonical genetic code was measured taking into account the harmful consequences resulting from point mutations leading to the replacement of one amino acid for another. There are two basic theories to measure the level of optimization: the statistical approach [2], which compares the canonical genetic code with many randomly generated alternative ones, and the engineering approach [3], which compares the canonical code with the best possible alternative.
Evolutionary Computing
We experimented with the use of evolutionary computing to search for better adapted hypothetical codes and as a method to guess the difficulty in finding such alternative codes, allowing to clearly situate the canonical code in the fitness landscape. This provides a new perspective in the open debate between the use of the statistical approach, which postulates that the genetic code conserves amino acid properties far better than expected from a random code, and the engineering approach, which tends to indicate that the canonical genetic code is still far from optimal. We used two models of hypothetical codes in our experiments: one that reflects the known examples of codon reassignment and the model most used in the two approaches which reflects the current genetic code translation table. Our results indicate that, although the standard code is far from a possible optimum considering both models, when the more realistic model of the codon reassignments was used, the evolutionary algorithm had more difficulty to overcome the efficiency of the canonical genetic code.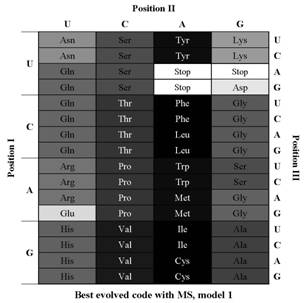 |
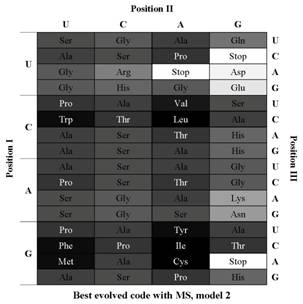 |
MS - Mean Square change in an amino acid property (polar requirement) resulting from all possible changes to each base of all the codons within a given code (it measures que optimality of a code).
tMS – MS that takes into account the mistranslation in each codon base position.
[1] Crick F: The origin of the genetic code. Journal of Theoretical Biology 1968, 38:367-379.
[2] Freeland SJ, Knight RD, Landweber LF: Measuring adaptation within the genetic code. Trends in Biochemical Sciences 2000, 25(2):44-45.
[3] Di Giulio M: The origin of the genetic code. Trends in Biochemical Sciences 2000, 25(2):44.

